Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Feflowlib: How to modify the project-file after converting a FEFLOW model.#
This example shows how to convert a FEFLOW model and how to modify the corresponding OGS project file and boundary meshes after conversion.
Necessary imports
import tempfile
import xml.etree.ElementTree as ET
from pathlib import Path
import ogstools as ot
from ogstools.examples import feflow_model_2D_CT_t_560
1. Load a FEFLOW model (.fem) as a FeflowModel object to further work it. During the initialisation, the FEFLOW file is converted.
temp_dir = Path(tempfile.mkdtemp("converted_models"))
feflow_model = ot.FeflowModel(feflow_model_2D_CT_t_560, temp_dir / "CT_model")
Get the mesh and run the FEFLOW model in OGS.
mesh = feflow_model.mesh
feflow_model.setup_prj(
end_time=1e11, time_stepping=[(1, 1e6), (1, 1e8), (1, 1e10)]
)
feflow_model.run()
Project file written to output.
Simulation: /tmp/tmpa9pfbe9nconverted_models/CT_model.prj
Status: finished successfully.
Execution took 0.8928711414337158 s
Plot the results.
ms = ot.MeshSeries(temp_dir / "CT_model.pvd")
ogs_sim_res = ms.mesh(ms.timesteps[-1])
ot.plot.contourf(ogs_sim_res, "single_species")
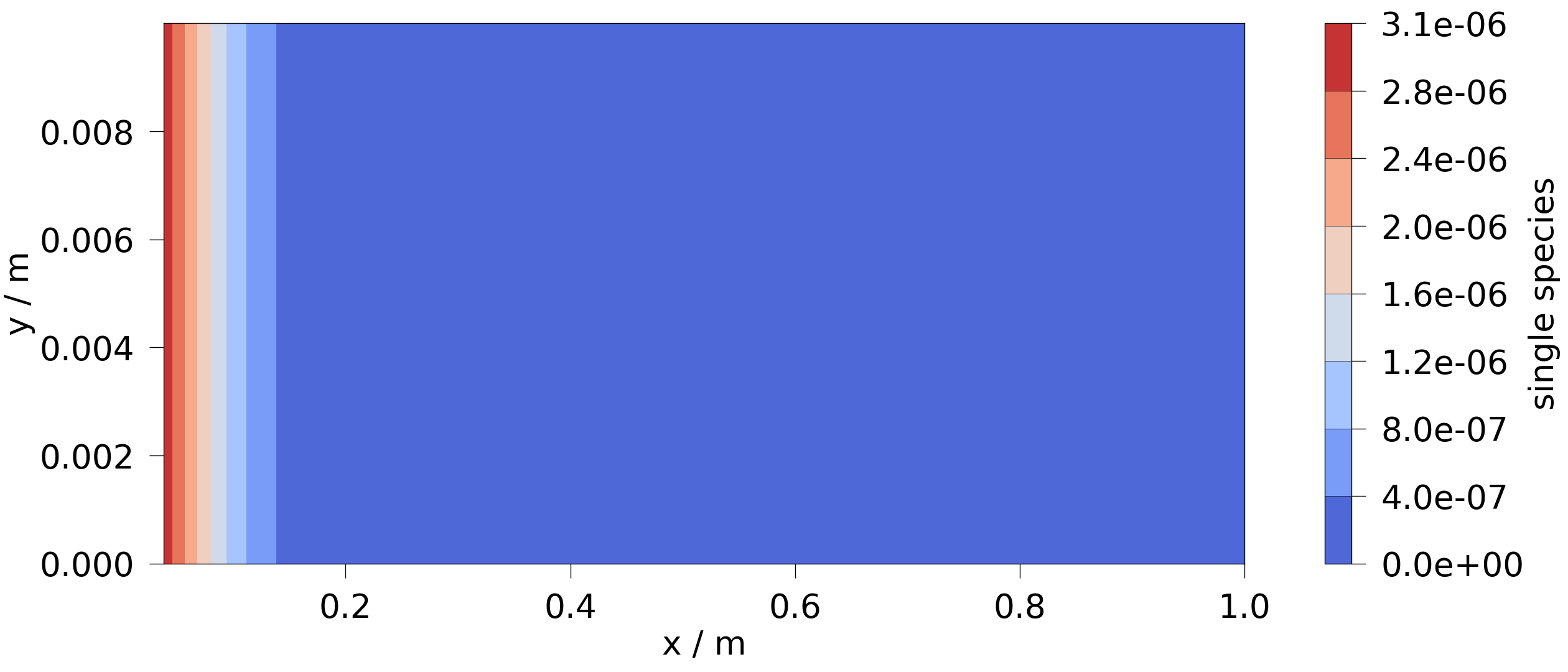
<Figure size 2520x1080 with 2 Axes>
4. Replace the scalar pore diffusion constant by a tensor to introducec anisotropy. How to manipulate a prj file also is explained in this example: # Run a simulation - from a Project file.
tensor = """
1e-9 1e-12
"""
feflow_model.project.replace_phase_property_value(
mediumid=0, component="single_species", name="pore_diffusion", value=tensor
)
feflow_model.save()
model_prjfile = ET.parse(temp_dir / "CT_model.prj")
ET.dump(model_prjfile)
<OpenGeoSysProject>
<meshes>
<mesh>CT_model.vtu</mesh>
<mesh>single_species_P_BC_MASS.vtu</mesh>
</meshes>
<processes>
<process>
<name>CT</name>
<type>ComponentTransport</type>
<coupling_scheme>staggered</coupling_scheme>
<integration_order>2</integration_order>
<specific_body_force>0 0</specific_body_force>
<secondary_variables>
<secondary_variable internal_name="darcy_velocity" output_name="v" />
</secondary_variables>
<process_variables>
<pressure>HEAD_OGS</pressure>
<concentration>single_species</concentration>
</process_variables>
</process>
</processes>
<media>
<medium id="0">
<phases>
<phase>
<type>AqueousLiquid</type>
<properties>
<property>
<name>viscosity</name>
<type>Constant</type>
<value>1</value>
</property>
<property>
<name>density</name>
<type>Constant</type>
<value>1</value>
</property>
</properties>
<components>
<component>
<name>single_species</name>
<properties>
<property>
<name>decay_rate</name>
<type>Constant</type>
<value>0.0</value>
</property>
<property>
<name>pore_diffusion</name>
<type>Constant</type>
<value>
1e-9 1e-12
</value>
</property>
<property>
<name>retardation_factor</name>
<type>Constant</type>
<value>16441.72737282367</value>
</property>
</properties>
</component>
</components>
</phase>
</phases>
<properties>
<property>
<name>porosity</name>
<type>Constant</type>
<value>0.10999999940395355</value>
</property>
<property>
<name>longitudinal_dispersivity</name>
<type>Constant</type>
<value>0.0</value>
</property>
<property>
<name>transversal_dispersivity</name>
<type>Constant</type>
<value>0.0</value>
</property>
<property>
<name>permeability</name>
<type>Constant</type>
<value>1.1574074074074073e-05</value>
</property>
</properties>
</medium>
</media>
<time_loop>
<processes>
<process ref="CT">
<nonlinear_solver>basic_picard</nonlinear_solver>
<convergence_criterion>
<type>DeltaX</type>
<norm_type>NORM2</norm_type>
<reltol>1e-6</reltol>
</convergence_criterion>
<time_discretization>
<type>BackwardEuler</type>
</time_discretization>
<time_stepping>
<type>FixedTimeStepping</type>
<t_initial>0</t_initial>
<t_end>100000000000.0</t_end>
<timesteps>
<pair>
<repeat>1</repeat>
<delta_t>1000000.0</delta_t>
</pair>
<pair>
<repeat>1</repeat>
<delta_t>100000000.0</delta_t>
</pair>
<pair>
<repeat>1</repeat>
<delta_t>10000000000.0</delta_t>
</pair>
</timesteps>
</time_stepping>
</process>
<process ref="CT">
<nonlinear_solver>basic_picard</nonlinear_solver>
<convergence_criterion>
<type>DeltaX</type>
<norm_type>NORM2</norm_type>
<reltol>1e-6</reltol>
</convergence_criterion>
<time_discretization>
<type>BackwardEuler</type>
</time_discretization>
<time_stepping>
<type>FixedTimeStepping</type>
<t_initial>0</t_initial>
<t_end>100000000000.0</t_end>
<timesteps>
<pair>
<repeat>1</repeat>
<delta_t>1000000.0</delta_t>
</pair>
<pair>
<repeat>1</repeat>
<delta_t>100000000.0</delta_t>
</pair>
<pair>
<repeat>1</repeat>
<delta_t>10000000000.0</delta_t>
</pair>
</timesteps>
</time_stepping>
</process>
</processes>
<output>
<type>VTK</type>
<prefix>/tmp/tmpa9pfbe9nconverted_models/CT_model</prefix>
<timesteps>
<pair>
<repeat>1</repeat>
<each_steps>1</each_steps>
</pair>
</timesteps>
<variables>
<variable>single_species</variable>
<variable>HEAD_OGS</variable>
</variables>
<fixed_output_times>100000000000.0</fixed_output_times>
</output>
<global_process_coupling>
<max_iter>1</max_iter>
<convergence_criteria>
<convergence_criterion>
<type>DeltaX</type>
<norm_type>NORM2</norm_type>
<reltol>1e-10</reltol>
</convergence_criterion>
<convergence_criterion>
<type>DeltaX</type>
<norm_type>NORM2</norm_type>
<reltol>1e-10</reltol>
</convergence_criterion>
</convergence_criteria>
</global_process_coupling>
</time_loop>
<parameters>
<parameter>
<name>C0</name>
<type>Constant</type>
<value>0</value>
</parameter>
<parameter>
<name>p0</name>
<type>Constant</type>
<value>0</value>
</parameter>
<parameter>
<name>single_species_P_BC_MASS</name>
<type>MeshNode</type>
<mesh>single_species_P_BC_MASS</mesh>
<field_name>single_species_P_BC_MASS</field_name>
</parameter>
</parameters>
<process_variables>
<process_variable>
<name>single_species</name>
<components>1</components>
<order>1</order>
<initial_condition>C0</initial_condition>
<boundary_conditions>
<boundary_condition>
<type>Dirichlet</type>
<mesh>single_species_P_BC_MASS</mesh>
<parameter>single_species_P_BC_MASS</parameter>
</boundary_condition>
</boundary_conditions>
</process_variable>
<process_variable>
<name>HEAD_OGS</name>
<components>1</components>
<order>1</order>
<initial_condition>p0</initial_condition>
</process_variable>
</process_variables>
<nonlinear_solvers>
<nonlinear_solver>
<name>basic_picard</name>
<type>Picard</type>
<max_iter>100</max_iter>
<linear_solver>general_linear_solver</linear_solver>
</nonlinear_solver>
</nonlinear_solvers>
<linear_solvers>
<linear_solver>
<name>general_linear_solver</name>
<lis>-i cg -p jacobi -tol 1e-10 -maxiter 100000</lis>
<eigen>
<solver_type>CG</solver_type>
<precon_type>DIAGONAL</precon_type>
<max_iteration_step>100000</max_iteration_step>
<error_tolerance>1e-10</error_tolerance>
</eigen>
</linear_solver>
</linear_solvers>
</OpenGeoSysProject>
Remove some points of the boundary mesh, which is part of the subdomains.
bounds = [0.037, 0.039, 0.003, 0.006, 0, 0]
new_bc_mesh = feflow_model.subdomains["single_species_P_BC_MASS"].clip_box(
bounds, invert=False
)
feflow_model.subdomains["single_species_P_BC_MASS"] = new_bc_mesh
Run the model.
feflow_model.run(overwrite=True)
ms = ot.MeshSeries(temp_dir / "CT_model.pvd")
ogs_sim_res = ms.mesh(ms.timesteps[-1])
ot.plot.contourf(ogs_sim_res, "single_species")
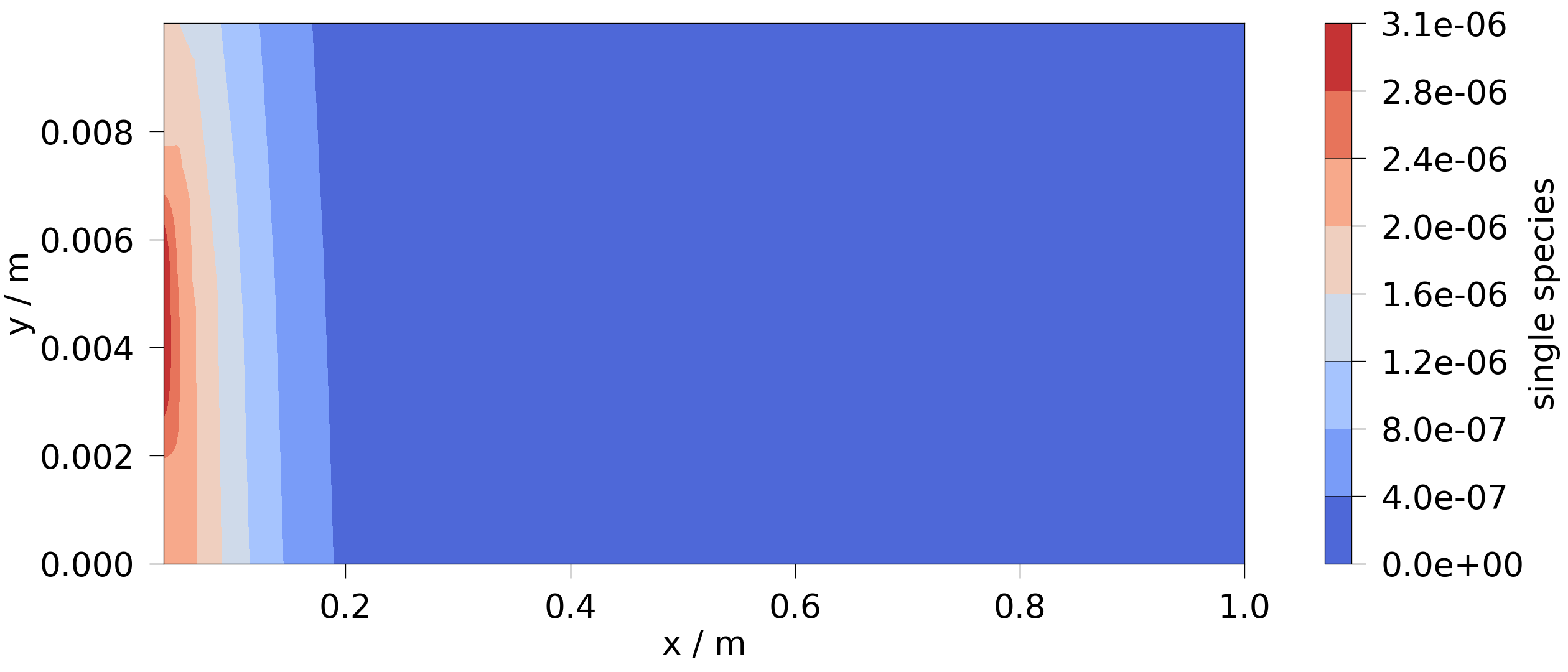
Project file written to output.
Simulation: /tmp/tmpa9pfbe9nconverted_models/CT_model.prj
Status: finished successfully.
Execution took 0.9962406158447266 s
<Figure size 2520x1080 with 2 Axes>
Total running time of the script: (0 minutes 3.576 seconds)

