Note
Go to the end to download the full example code. or to run this example in your browser via Binder
How to Create Time Slices#
In this example we show how to create a filled contourplot of transient data over a sampling line. For this purpose we use a component transport example from the ogs benchmark gallery (https://www.opengeosys.org/docs/benchmarks/hydro-component/elder/).
To see this benchmark results over all timesteps have a look at How to create Animations.
Let’s load the data which we want to investigate.
import numpy as np
import ogstools as ot
from ogstools import examples
mesh_series = examples.load_meshseries_CT_2D_XDMF().scale(time=("s", "a"))
si = ot.variables.saturation
Now we setup two sampling lines.
pts_vert = np.linspace([25, 0, -75], [25, 0, 75], num=300)
pts_diag = np.linspace([25, 0, 75], [100, 0, 0], num=300)
fig = mesh_series.mesh(-1).plot_contourf(si, vmin=0)
fig.axes[0].plot(pts_vert[:, 0], pts_vert[:, 2], "-k", linewidth=3)
fig.axes[0].plot(pts_diag[:, 0], pts_diag[:, 2], "-.k", linewidth=3)
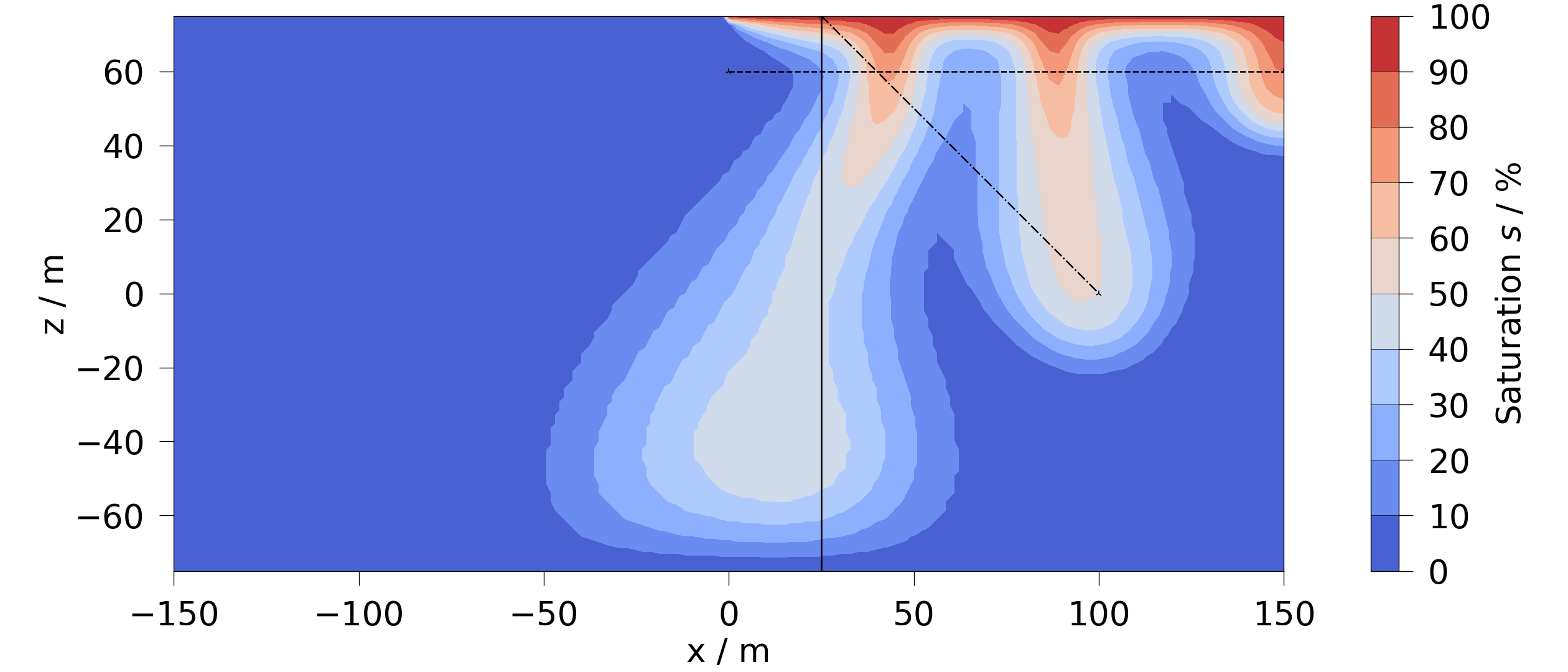
[<matplotlib.lines.Line2D object at 0x792a997b2b00>]
Here, we first show a regular line sample plot for the vertical sampling line for each timestep.
ms_vert = ot.MeshSeries.extract_probe(mesh_series, pts_vert)
labels = [f"{tv:.1f} a" for tv in ms_vert.timevalues]
fig = ot.plot.line(ms_vert, si, "z", labels=labels, colors="coolwarm")
fig.tight_layout()
As the above kind of plot is getting cluttered for lots of timesteps we
provide a function to create a filled contour plot over the transient data.
The function plot_time_slice()
creates a heatmap over time and space.
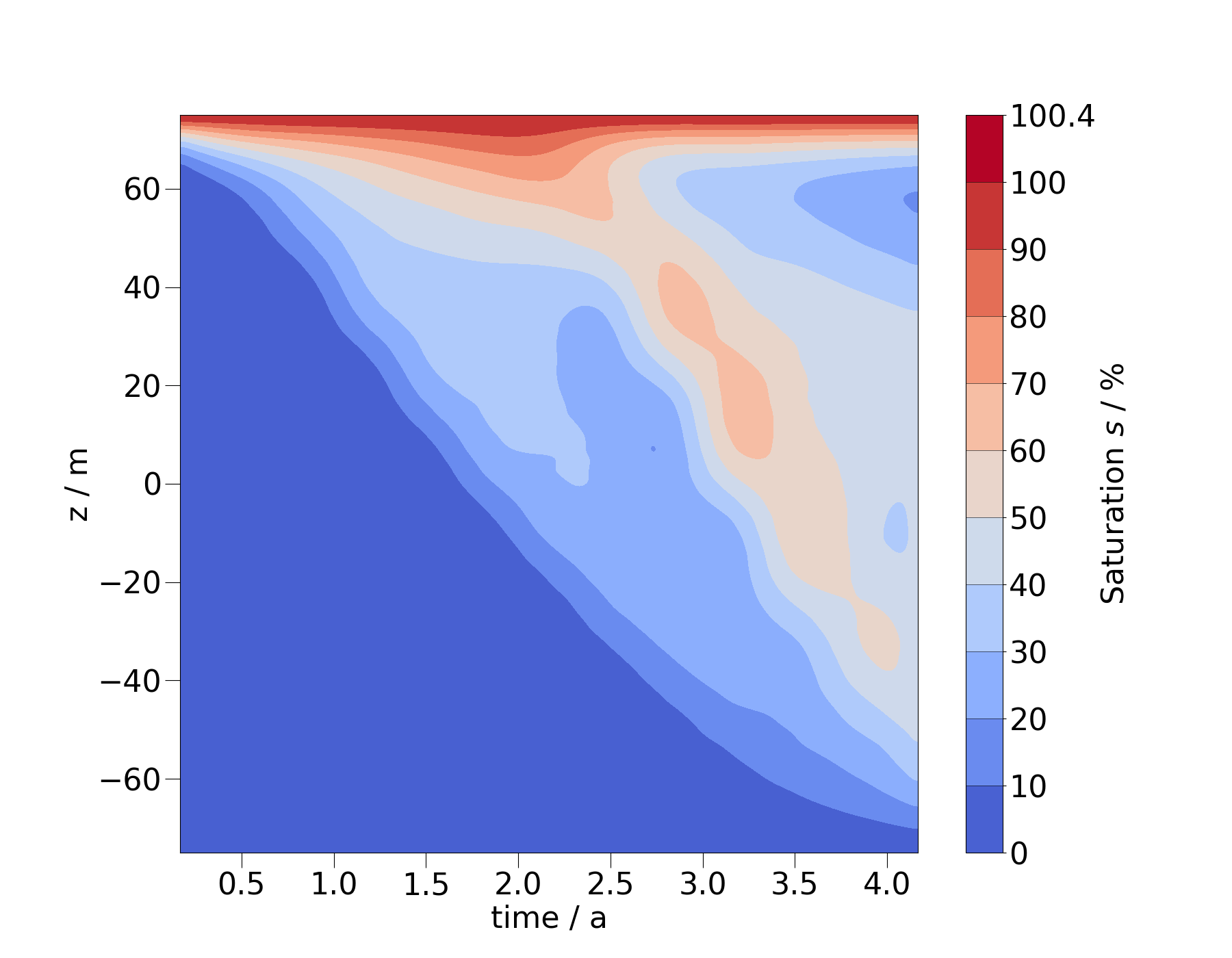
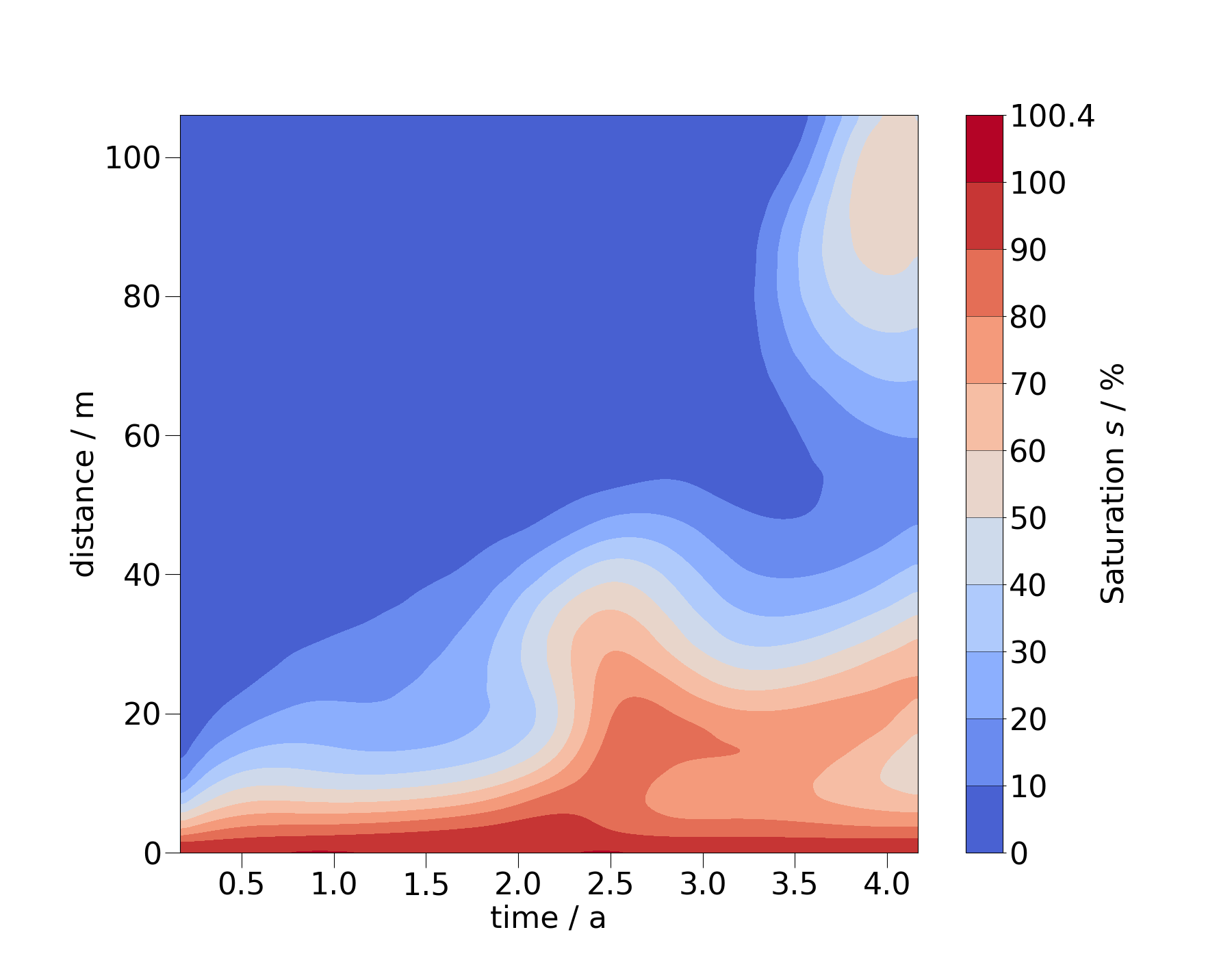
fig = ms_vert.plot_time_slice("time", "z", si, vmin=0, vmax=100)
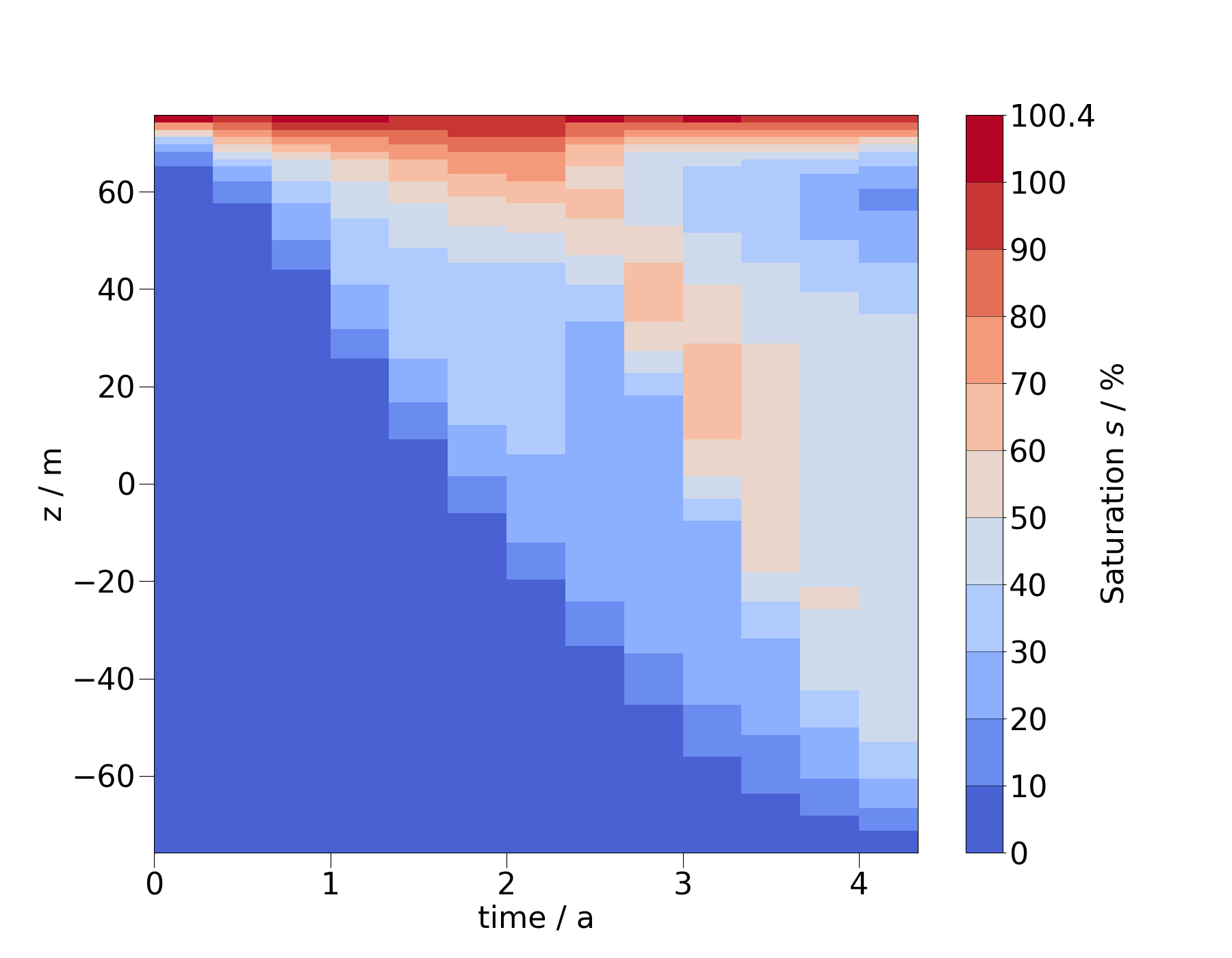
The stepping in this heatmap corresponds to the individual timesteps. To create a smoother image, we can resample the MeshSeries to more timesteps.
ms_vert_fine = ot.MeshSeries.resample(ms_vert, np.linspace(0, 4.2, 300))
fig = ms_vert_fine.plot_time_slice("time", "z", si, vmin=0, vmax=100)
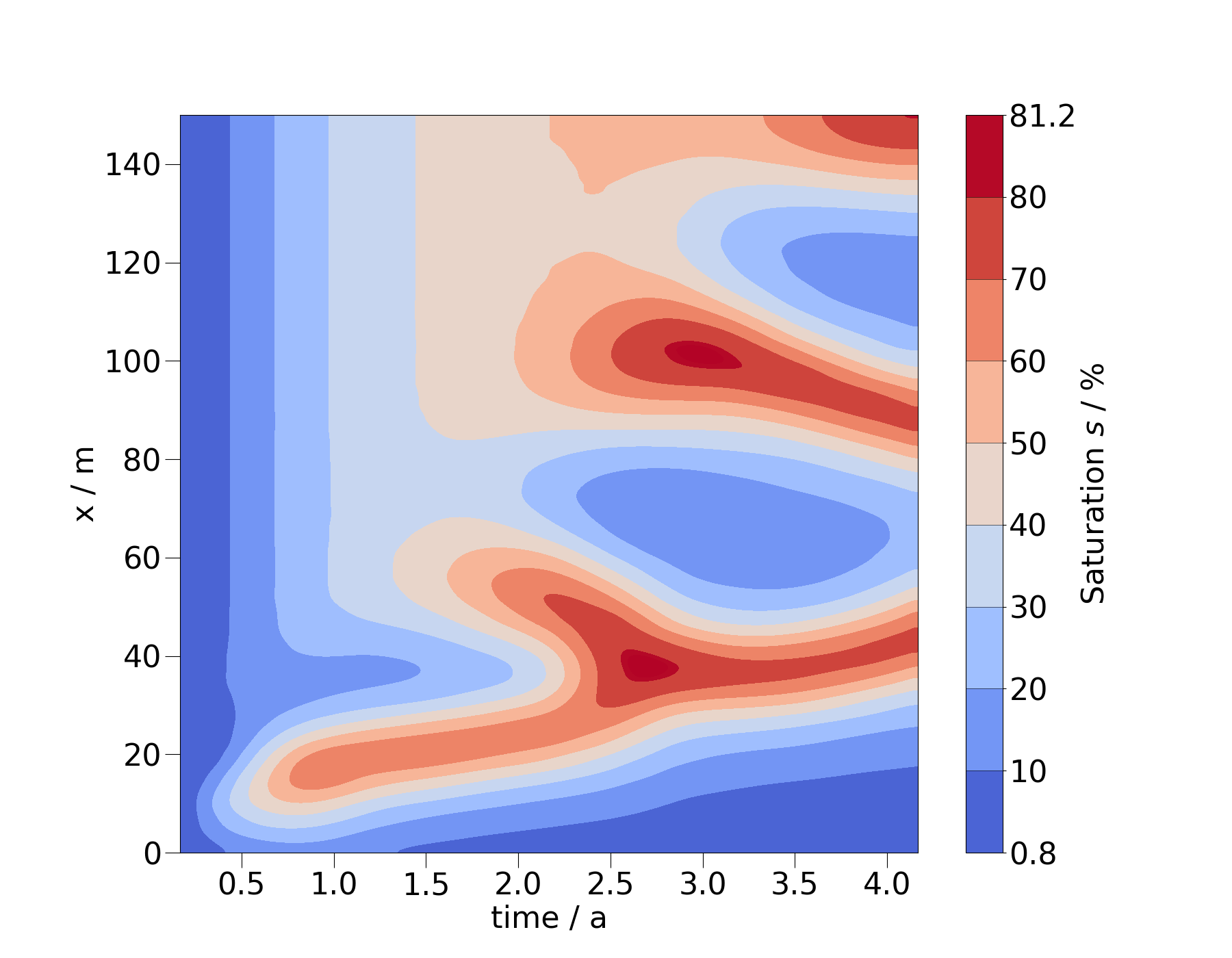
You can also change the order of the arguments for spatial coordinate and time to flip the x- and y-axis.
ms_diag = ot.MeshSeries.extract_probe(mesh_series, pts_diag)
ms_diag_fine = ot.MeshSeries.resample(ms_diag, np.linspace(0, 4.2, 300))
fig = ms_diag_fine.plot_time_slice("x", "time", si, vmin=0, vmax=100)
fig.axes[0].invert_yaxis()
Total running time of the script: (0 minutes 4.597 seconds)

