Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Read mesh from file (vtu or xdmf) into pyvista mesh#
import numpy as np
from ogstools import examples
To read your own data as a MeshSeries you can do:
from ogstools.meshlib import MeshSeries
mesh_series = MeshSeries("filepath/filename_pvd_or_xdmf")
MeshSeries takes as mandatory argument a str OR pathlib.Path that represents the location of the pvd or xdmf file. Here, we load example data and print the meta information:
ms = examples.load_meshseries_HT_2D_XDMF()
ms
<ogstools.meshlib.mesh_series.MeshSeries object at 0x792aa87fea70>
Accessing time values#
Time values (and spatial coordinates) can be unit transformed via
scale(). Either pass a tuple
to convert from the first to the second unit or pass a scaling factor.
print(f"First 3 time values are: {ms.timevalues[:3]} s.")
ms = ms.scale(time=("s", "h"))
print(f"Last time value is: {ms.timevalues[-1]} h.")
ms = ms.scale(time=3600.0)
print(f"Last time value is: {ms.timevalues[-1]} s.")
First 3 time values are: [ 0. 900. 1800.] s.
Last time value is: 24.0 h.
Last time value is: 86400.0 s.
Accessing meshes#
To get a single mesh at a specified timestep you can use the mesh() method of a MeshSeries object (in this example: ms). Another way to to do so is by indexing the MeshSeries object with brackets (ms[timestep]). Besides custom ogstools functions you can use all available pyvista functions. Here we use pyvista’s plot function.
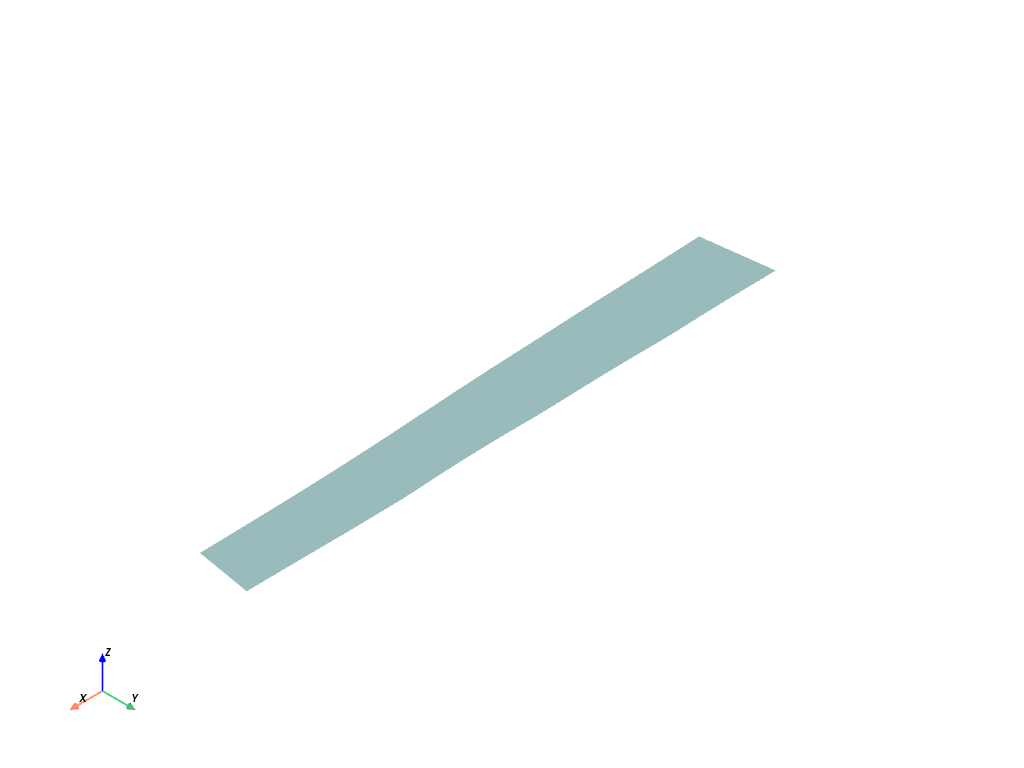
mesh_ts10 = ms.mesh(10) # or ms[10]
mesh_ts10.plot(show_edges=True)
Indexing#
MeshSeries.values(“<variable_name>”)` returns a numpy ndarray. It allows multidimensional indexing on ndarrays.
Typically, the first dimension is the time step, second dimension is the number of points/cells, and the last dimension is the number of components of the variable.
By default, values would read the entire dataset. If only a subset of the
MeshSeries should be read you can select the relevant timesteps by indexing /
slicing the MeshSeries directly. This selection will also be adhered to if you
read individual meshes.
ms = examples.load_meshseries_HT_2D_XDMF()
# Temperature is a scalar, Darcy velocity is a vector with 2 components.
# Both are defined at points.
print("Entire dataset:", np.shape(ms.values("temperature")))
print("Every second timestep:", np.shape(ms[::2].values("temperature")))
print("Last two steps:", np.shape(ms[-2:].values("darcy_velocity")))
Entire dataset: (97, 190)
Every second timestep: (49, 190)
Last two steps: (2, 190, 2)
To select points or cells you can use the extract method to specify the
corresponding ids.
temp_at_points = ms.extract([2, 3, 4]).values("temperature")
print("Data on extracted points:", np.shape(temp_at_points))
print("Temperatures at last timestep:", temp_at_points[-1])
Data on extracted points: (97, 3)
Temperatures at last timestep: [352.9994477 303. 353.00000001]
You can also use pyvista dataset filters to transform the domain for the
entire MeshSeries.
ms_right_half = ms.transform(
lambda mesh: mesh.clip("x", mesh.center, crinkle=True)
)
temp_right_half = ms_right_half.values("temperature")
print("Data on clipped domain:", np.shape(temp_right_half))
Data on clipped domain: (97, 114)
Let’s plot the last timestep of the transformed MeshSeries.
fig = ms_right_half[-1].plot_contourf("temperature")
Total running time of the script: (0 minutes 2.787 seconds)

