Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Spatial & temporal refinement - nuclear decay#
This example shows one possible implementation of how to do a convergence study
with spatial and temporal refinement. For this, a simple model using a time
dependent heat source on one side and constant temperature on the opposite side
was set up. The heat source is generated with the
ogstools.physics.nuclearwasteheat model.
Here is some theoretical background for the topic of grid convergence:
At least three meshes from simulations of increasing refinement are required for the convergence study. The third finest mesh is chosen per default as the topology to evaluate the results on.
The results to analyze are generated on the fly with the following code. If you are only interested in the convergence study, please skip to Temperature convergence at maximum heat production (t=30 yrs).
First, the required packages are imported and a temporary output directory is created:
from pathlib import Path
from tempfile import mkdtemp
import matplotlib.pyplot as plt
import numpy as np
from IPython.display import HTML
from scipy.constants import Julian_year as sec_per_yr
import ogstools as ot
from ogstools import examples, physics, studies, workflow
temp_path = Path(mkdtemp(prefix="nuclear_decay"))
Let’s run the different simulations with increasingly fine spatial and
temporal discretization via ogs6py. The mesh and its boundaries are generated
easily via gmsh and Meshes. from_gmsh().
First some definitions:
n_refinements = 4
time_step_sizes = [30.0 / (2.0**r) for r in range(n_refinements)]
prefix = "stepsize_{0}"
sim_results = []
msh_path = temp_path / "rect.msh"
script_path = Path(examples.pybc_nuclear_decay).parent
prj_path = examples.prj_nuclear_decay
edge_cells = [5 * 2**i for i in range(n_refinements)]
Now the actual simulations:
for dt, n_cells in zip(time_step_sizes, edge_cells, strict=False):
ot.meshlib.rect(lengths=100.0, n_edge_cells=(n_cells, 1), out_name=msh_path)
meshes = ot.Meshes.from_gmsh(msh_path, log=False)
mesh_path = temp_path / f"dt_{dt}_n_cells_{n_cells}"
mesh_path.mkdir(parents=True, exist_ok=True)
meshes.save(mesh_path)
prj = ot.Project(output_file=temp_path / "default.prj", input_file=prj_path)
prj.replace_text(str(dt * sec_per_yr), ".//delta_t")
prj.replace_text(prefix.format(dt), ".//prefix")
prj.write_input()
ogs_args = f"-m {mesh_path} -o {temp_path} -s {script_path}"
prj.run_model(write_logs=False, args=ogs_args)
sim_results += [temp_path / (prefix.format(dt) + "_domain.xdmf")]
Project file written to output.
Simulation: /tmp/nuclear_decayj8est85z/default.prj
Status: finished successfully.
Execution took 8.482245922088623 s
Project file written to output.
Simulation: /tmp/nuclear_decayj8est85z/default.prj
Status: finished successfully.
Execution took 7.988728046417236 s
Project file written to output.
Simulation: /tmp/nuclear_decayj8est85z/default.prj
Status: finished successfully.
Execution took 8.960731744766235 s
Project file written to output.
Simulation: /tmp/nuclear_decayj8est85z/default.prj
Status: finished successfully.
Execution took 8.95310115814209 s
Let’s extract the temperature evolution and the applied heat via vtuIO and plot both:
time = np.append(0.0, np.geomspace(1.0, 180.0, num=100))
repo = physics.nuclearwasteheat.repo_2020_conservative
heat = repo.heat(time, time_unit="yrs", power_unit="kW")
fig, (ax1, ax2) = plt.subplots(figsize=(8, 8), nrows=2, sharex=True)
ax2.plot(time, heat, lw=2, label="reference", color="k")
for sim_result, dt in zip(sim_results, time_step_sizes, strict=False):
mesh_series = ot.MeshSeries(sim_result).scale(time=("s", "yrs"))
results = {"heat_flux": [], "temperature": []}
for ts in mesh_series.timesteps:
mesh = mesh_series.mesh(ts)
results["temperature"] += [np.max(mesh.point_data["temperature"])]
max_T = ot.variables.temperature.transform(results["temperature"])
# times 2 due to symmetry, area of repo, to kW
results["heat_flux"] += [np.max(mesh.point_data["heat_flux"][:, 0])]
tv = np.asarray(mesh_series.timevalues)
ax1.plot(tv, max_T, lw=1.5, label=f"{dt=}")
edges = np.append(0, tv)
mean_t = 0.5 * (edges[1:] + edges[:-1])
applied_heat = repo.heat(mean_t, time_unit="yrs", power_unit="kW")
ax2.stairs(applied_heat, edges, lw=1.5, label=f"{dt=}", baseline=None)
ax2.set_xlabel("time / yrs")
ax1.set_ylabel("max T / °C")
ax2.set_ylabel("heat / kW")
ax1.legend()
ax2.legend()
fig.show()
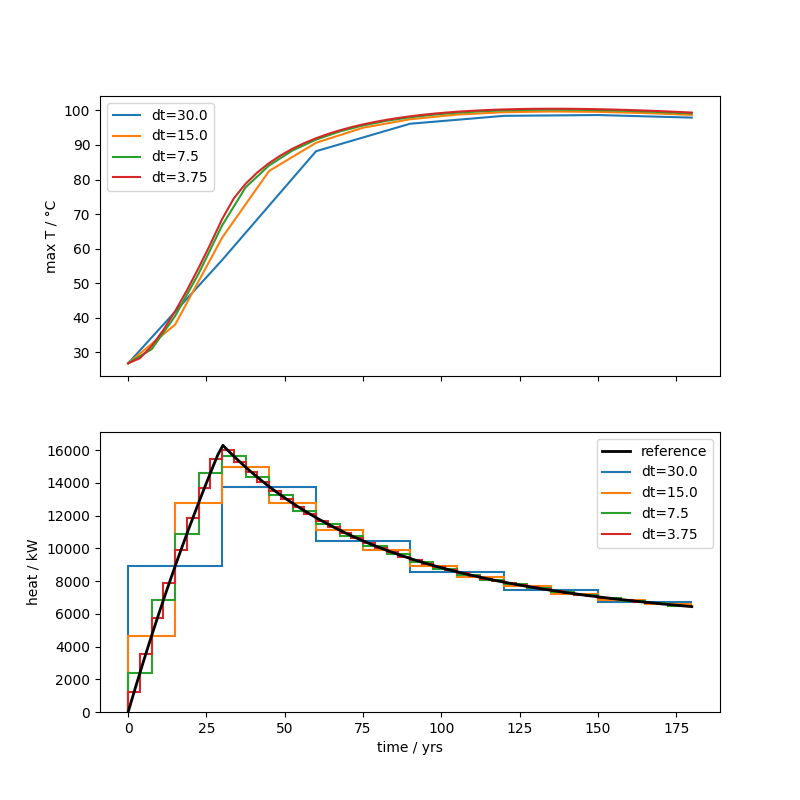
Temperature convergence at maximum heat production (t=30 yrs)#
The grid convergence at this timepoint deviates significantly from 1, meaning the convergence is suboptimal (at least on the left boundary where the heating happens). The chosen timesteps are still to coarse to reach an asymptotic range of convergence. The model behavior at this early part of the simulation is still very dynamic and needs finer timesteps to be captured with great accuracy. Nevertheless, the maximum temperature converges quadratically, as expected.
report_name = temp_path / "report.ipynb"
studies.convergence.run_convergence_study(
output_name=report_name,
mesh_paths=sim_results,
timevalue=30 * sec_per_yr,
variable_name="temperature",
refinement_ratio=2.0,
)
HTML(workflow.jupyter_to_html(report_name, show_input=False))
Temperature convergence at maximum temperature (t=150 yrs)#
The temperature convergence at this timevalue is much closer to 1, indicating a better convergence behaviour, which is due to the temperature gradient now changing only slowly. Convergence order is again quadratic.
studies.convergence.run_convergence_study(
output_name=report_name,
mesh_paths=sim_results,
timevalue=150 * sec_per_yr,
variable_name="temperature",
refinement_ratio=2.0,
)
HTML(workflow.jupyter_to_html(report_name, show_input=False))
Convergence evolution over all timesteps#
We can also run the convergence evaluation on all timesteps and look at the relative errors (between finest discretization and Richardson extrapolation) and the convergence order over time to get a better picture of the transient model behavior.
mesh_series = [ot.MeshSeries(sim_result) for sim_result in sim_results]
evolution_metrics = studies.convergence.convergence_metrics_evolution(
mesh_series, ot.variables.temperature, units=["s", "yrs"]
)
0%| | 0/7 [00:00<?, ?it/s]
43%|████▎ | 3/7 [00:00<00:00, 21.42it/s]
86%|████████▌ | 6/7 [00:00<00:00, 20.07it/s]
100%|██████████| 7/7 [00:00<00:00, 20.10it/s]
Looking at the errors, we see a higher error right at the beginning. This is likely due to the more dynamic behavior at the beginning.
fig = studies.convergence.plot_convergence_error_evolution(
evolution_metrics, error_type="absolute"
)
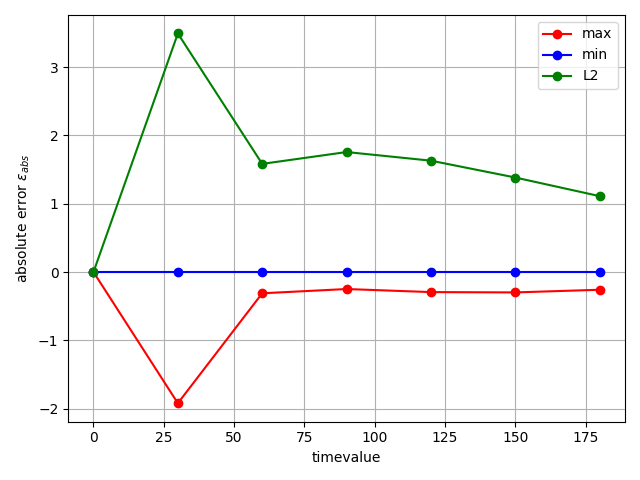
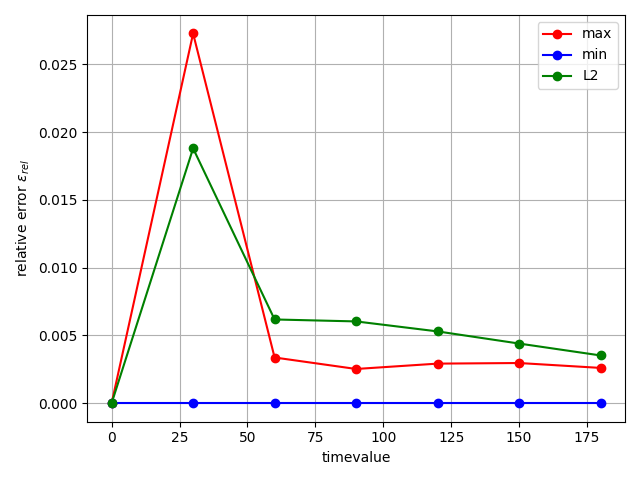
fig = studies.convergence.plot_convergence_error_evolution(
evolution_metrics, error_type="relative"
)
A look at the convergence order evolution shows almost quadratic convergence over the whole timeframe. For the maximum temperature we even get better than quadratic behavior, which is coincidentally and most likely model dependent.
fig = studies.convergence.plot_convergence_order_evolution(evolution_metrics)
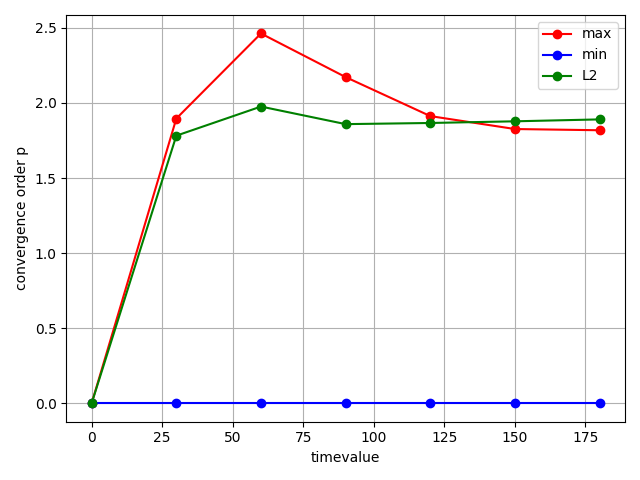
Total running time of the script: (1 minutes 2.891 seconds)

